ICBHI 2022
Menu

Welcome to the fourth edition of the Scientific Challenge Competition of the Health Informatics Working Group of the IFMBE.
In this edition of the IFMBE Scientific Challenge, participants are asked to identify individual heart beats from ballistocardiogram (BCG) records of patients and healthy volunteers. The BCG signal is obtained from an electromechanical film (EMFi) attached to the seat of a chair. An electrocardiogram (ECG) signal was acquired simultaneously for reference.

Figure 1: ECG and BCG signals. IJK wave always lags its corresponding QRS wave.
The acquisition of signals from unobtrusive sensors based on EMFi or other technologies would simplify patient care by enabling pervasive or ambient monitoring. These sensors can be installed in chairs, couches, and beds to obtain data without interfering with the patient’s daily activities. Also, their use in emergency waiting rooms would allow on-line triage and more information for clinicians before their physical interaction.
The difficulty in detecting heart beats from BCG comes from the fact that BCG is a very weak signal, produced by the mechanical movement resulting from the heart systolic contraction. The acquisition must have a high sensibility, which makes it more vulnerable to noise. Talking, adjusting on the seat or even people walking in the vicinity can introduce noise in the BCG signal. In this challenge, we invite you to design and test new algorithms for BCG processing. The accurate detection of individual beats can lead to more advanced monitoring, such as arrythmia detection in patients with atrial fibrillation, a commonly underdiagnosed condition that leads in time to more serious health complications.
The database is composed of 4 datasets from different experiments, coming from 2 groups of people: healthy volunteers and patients. The patients were previously diagnosed with AF, waiting for their routine check-up at our local hospital. In the experiment, the subjects were sitting in an EMFi-fitted chair for measuring BCG and wearing 3 ECG electrodes for a 1-derivation ECG record. Both signals were acquired simultaneously. The experiment protocol was approved by the necessary ethics committee and all volunteers agreed to an informed consent. ECG beats were reviewed by a cardiologist and are included for reference in the public dataset.
In order to participate as an individual or a collective team you should first register to the Challenge in order to be able to access the public annotated dataset of ECG and BCG made available for the challenge and to submit your entries to the challenge.
For those wishing to compete officially, please follow the additional four steps described in the Rules and Deadlines section.
We expect the submission of an overview paper with the best solutions to a prestigious Scientific Journal, co-authored by participants of the challenge and considering the publications and discussions at ICBHI’22.
In this challenge, we adopt the general rules of the Physionet Competition. The challenge is structured in two phases: the unofficial (Phase I) and the official (Phase II) phases. Participants may submit up to 15 entries over both the unofficial and official phases of the competition (see Table 1). Each participant may receive scores for up to five entries submitted during the unofficial phase and ten entries during the official phase. Unused entries may not be carried over to later phases. Entries that cannot be scored (because of missing components, improper formatting, or excessive run time) are not counted against the entry limits.
All deadlines occur at noon GMT (UTC) on the dates mentioned below.
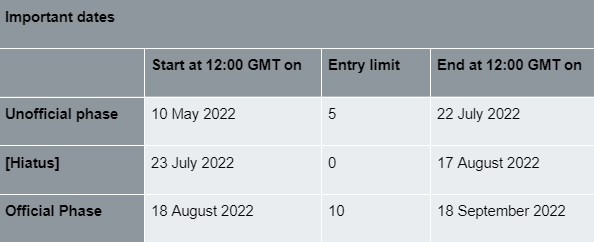
All official entries must be received no later than 12:00 GMT on the 18 September 2022. In the interest of fairness to all participants, late entries will not be accepted or scored.
To be eligible you must do all of the following:
Please do not submit analysis of this year’s Scientific Challenge data to other conferences or journals until after ICBHI’22 has taken place, so the competitors are able to discuss the results in a single forum. We expect the submission of an overview paper with the best solutions to a prestigious Scientific Journal, co-authored by participants of the challenge and taking into account the publications and discussions at ICBHI’22.
In case all the necessary files are included in your entry, and they run as expected, your entry is evaluated and you will receive your provisional scores when the test is complete. An automatic email will be delivered informing you of the results. The evaluation will be made on a weekly basis. If you receive an error message instead, read it carefully and correct the problem(s) before resubmitting.
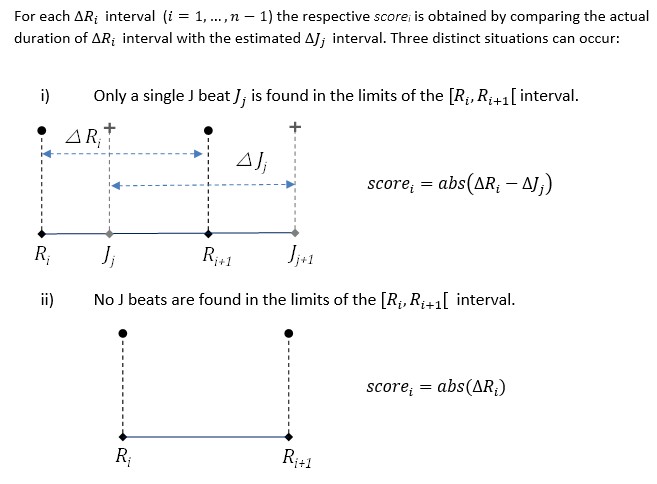
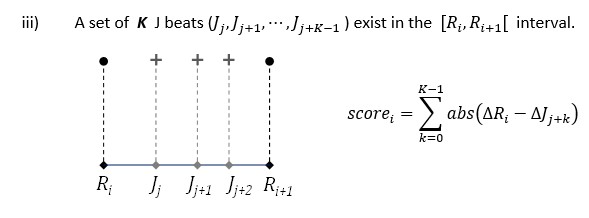
The overall score for your entry is computed based on the number of detected beat intervals.

a) Datasets
The dataset is divided in two parts: a public/training set and a private/testing set. The public partition of the database contains 83 records of different length and origin. The test set is unavailable to the public and will remain private for the purpose of scoring. The ECG with annotated beats is provided as a reference only. The algorithm has to be able to process the BCG signal alone (single channel), without references. Healthy volunteers and patients include both female and male subjects, of different weights and ages. The training and test sets have each been divided so that they are two sets of mutually exclusive populations (i.e., no recordings from the same subject/patient are in both training and test sets).
In the second phase, additional records will be made available in the public dataset.
Please note that many recordings are corrupted by various noise sources, such as talking, adjusting position, and moving in general.
Table 1: Different datasets used for the challenge.
| Dataset | Signals | Signal length | Comments |
| A01 | ECG, BCG | 13.5 min | Healthy volunteers with 2 records each: normal activities and after exercise. |
| A02 | ECG, BCG | 3 min | Patients previously diagnosed with AF (not necessarily presenting AF at the time of measurement) |
| A03 | ECG, BCG | 2 min | Healthy volunteers |
| A04 | ECG, BCG | 1 min | Healthy volunteers |
b) Input/output formats
To enter the ICBHI challenge, the participants have to develop an algorithm that is able to read a single BCG signal, providing as output the beat times in ms corresponding to the J point in the IJK wave, as shown in Figure 1, without any user interaction. The algorithm must be implemented in MATLAB or Python 3.x. Each team must upload to the ICBHI challenge website a zip, 7z, rar or tar.gz archive containing the following files:
We will run your code on the testing set and compare the contents of the results_RecordName.txt files that are generated with the ground-truth annotations of the health professionals. If everything runs as expected, you will get an email with the score of your submission.
In Phase I (Unofficial) your submissions will be tested on a subset of the testing set, while in Phase II (Official) they will be tested on the whole testing set. Thus, the grade for a specific algorithm may be different in each phase.
You can download the public set, the expected submission files and a sample challenge.m and challenge.py as reference with required inputs and outputs as zip files.